Breeding Value Prediction for Production Traits in Layers Using High-Density SNP Markers
Use of these markers increased accuracy up to two-fold for early selection and by up to 88 per cent for late selection, according to Anna Wolc and co-workers in Iowa State University's Animal Industry Report 2011.Summary and Implications
Accuracy of breeding values estimated by different methods using pedigree and high-density SNP genotypes in predicting the next generation in a commercial layer breeding line was evaluated. Early and late selection was considered. Use of markers increased accuracies up to two-fold for early selection and by up to 88 per cent for late selection.
Introduction
Through the application of genomic or whole-genome selection (Meuwissen et al. 2001), marker information from high-density SNP genotyping can improve accuracy of selection at young ages, shorten generation intervals and provide better control of inbreeding, which should lead to higher genetic gain per time unit. Multiple simulation studies have been conducted showing that the benefits of the technology depend on heritability, number and effects of QTL, population structure, size of the training data set, and other factors (Goddard, 2009). There are however few studies on real data. The objective of this study was to evaluate the accuracy of breeding values estimated using high-density SNP genotypes in predicting the next generation in a commercial layer breeding line, and to compare the accuracy of different methods of breeding value estimation.
Materials and Methods
The analyzed traits were: egg production (ePD, lPD), egg weight (eE3, eEW, lEW), egg color (eC3, eCO, lCO), shell strength (ePS, lPS), age at sexual maturity (eSM), body weight (lBW), albumen height (eAH, lAH), and yolk weight (eYW, lYW). With early (e) records taken at 26-28 wk and late (l) records at 42-46 wk of age. Predictions appropriate for early or late selection were compared. A total of 2,708 birds were genotyped for 23,356 segregating SNPs across the genome, including 1,563 females with records. Phenotypes on relatives without genotypes were also incorporated in the analysis (in total 13,049 production records). The data were analyzed using a Reduced Animal Model with a relationship matrix based on pedigree or on marker genotypes and with a Bayesian genomic selection method using model averaging. The methods were compared by using results to estimate breeding values in a validation set that consisted of individuals from the generation following training and evaluating the accuracy of the EBV based on the correlation of EBV with phenotypes corrected for fixed effects divided by the square root of heritability.
Results and Discussion
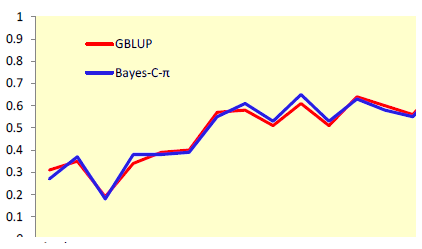
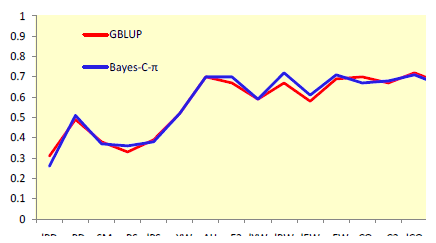
Use of markers increased accuracies up to two-fold for selection at an early age (Figure 1) and by up to 88% for selection at a later age (Figure 2). Improvements in accuracy at an early age could be attributed mostly to better estimates of parental EBV for shell quality and egg production, while for other egg quality traits it was mostly attributed to better estimates of Mendelian sampling effects. A relatively small number of markers were sufficient to explain most genetic variation for egg weight and body weight.
Further Reading
| - | You can view other reports in Iowa State University's Animal Industry Report 2011 by clicking here. |
April 2011