



Accuracy of Genomic EBV Using an Evenly Spaced, Low-Density SNP Panel in Broiler Chickens
Genomic selection using evenly-spaced low-density marker panels is a cost-effective choice for implementation of genomic selection, according to Chunkao Wang and co-workers in Iowa State University's Animal Industry Report 2011.Summary and Implications
Whole-genome or genomic selection is based on associations of large number of markers across the genome with phenotype but will require use of small SNP panels to be cost effective in chickens. The potential loss of accuracy of genotyping selection candidates with an evenly-spaced low-density marker panel and imputation of high-density SNP genotypes was evaluated in a commercial broiler chicken line.
Several methods were used to estimate marker effects. The loss in accuracy was less than five per cent for different methods and traits. Thus, genomic selection using evenly-spaced low-density marker panels is a cost-effective choice for implementation of genomic selection.
Introduction
Genomic selection (GS) using high-density (HD) marker panels provides opportunities to enhance genetic improvement of livestock but may not be cost-effective, especially for breeding programmes involving large numbers of selection candidates, due to the high costs of HD-SNP genotyping.
Previous research in our group has, however, shown that GS can be implemented by genotyping selection candidates for panels representing a subset of less than 400 of the HD markers well distributed across the genome, followed by imputing HD marker genotypes that had been observed on the parents and grandparents.
A key concern for this approach is potential loss of accuracy, which was evaluated for two traits in a commercial broiler breeding line.
Materials and Methods
The HD and evenly-spaced low density (ELD) panels had 36,455 and 384 SNPs and were genotyped using Illumina Infinium and KASPar Kbioscience platforms, respectively.
A total of 1,091 birds from three generations were genotyped with the HD panel as training data and 168 birds were genotyped with both HD and LD panels as validation data. The training data included the parents of the 168 birds in the validation data set. A fast rule-based method was used to infer SNP haplotypes of training-set individuals. A Gibbs sampler with overlapping blocks was used to estimate joint probabilities of allele segregation indicators at adjacent ELD SNPs for the validation individuals, utilising the haplotype information at ELD SNPs from training individuals. HD haplotypes in the training data and segregation probabilities at ELD SNPs for validation individuals were then used to estimate genotype probabilities of the missing HD SNPs in the validation individuals.
Genomic selection methods Bayes-A, -B (pi = 0.99), and -C (pi = 0.99) and GLUP were used to estimate marker effects in the training data for two traits: body weight and hen house production. Resulting estimates were used to estimate genomic breeding values of validation data using either their observed HD genotypes or their imputed HD genotypes from the ELD panel.
Using EBV computed from observed HD genotypes as gold standard, the loss in accuracy from using imputed genotypes was evaluated based on the difference in the correlation between EBV from observed and imputed HD genotypes.
Results and Discussion
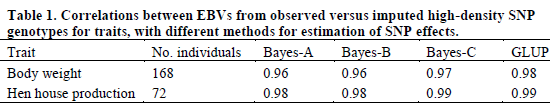
Correlations between EBVs from observed versus imputed HD genotypes are in Table 1. All correlations were greater than 0.95, which indicates that loss in accuracy from using the ELD panel is less than five per cent. Correlations were slightly smaller for body weight than for hen house production for all methods. Correlations were highest for GLUP.
Further Reading
| - | You can view other reports in Iowa State University's Animal Industry Report 2011 by clicking here. |
April 2011








